So what is this RData file format? It is a binary format and not so easy to inspect, but there is an option to save a file in ASCII:
So how does this file look like? Here is an annotated listing:
RDA2 Header: file type
A Ascii format
2 Format version 2
197123 R version information
131840 more R version information
1026 LISTSXP object: whole thing is packaged in a dotted pair list
1 SYMSXP object: symbol
262153 CHARSXP object: string
4 Length of string
ivec String: symbol name
13 INTSXP: integer vector
3 Length of integer vector
1 First element
2 Second element
3 Third element
254 NILVALUESXP: end of information
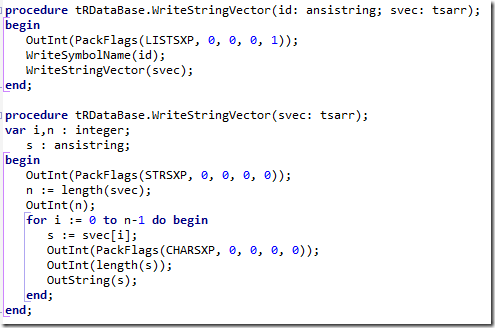
Using this information we could re-engineer writing R objects to an RData file. E.g. writing a string vector looks like:
(The tRDataBase name reflects this is a base class; we derive tRDataAscii, tRDataBinary and tRDataNetwork from this).
When we save objects without the “ascii=TRUE” flag, basically a compressed binary network format is used. The idea behind a network format is to write all binary data in a standardized big endian network byte ordering. This will allow a binary file written on one machine (e.g. with an Intel architecture) to be read on a different machine (actually there are not that many big-endian computer architectures left). This whole thing is then compressed using gzip.
Using an RDB2 header I can write a pure native binary format (that is without reordering bytes to a network byte ordering). It looks like R has decided not to support this format anymore:
So binary files always use the network byte ordering and have an RDX2 header.
Notes
- The load() function works perfectly fine with remote Rdata files:
> load(url("http://www.amsterdamoptimization.com/downloads/rvec.rdata"),verbose=T) Loading objects: x
- The goal of this exercise is to be able to generate .Rdata data sets from other environments. We don’t use R itself for this but rather write .Rdata files directly. Another approach would be to launch R, import the data set (e.g. using a CSV file) and then call save() to generate the .Rdata file. When doing this from a different programming language, it is possible to automate this using the R.dll. This is in fact how this interface in F# works (same thing for the rpy2 Python interface). In my setup I don’t need an R DLL and write .Rdata directly from the Delphi and C programming languages. So it is a little bit more lightweight and also does not require an installed R system.
- It is time for RData files to become the standard for Data Transfer.
- R Internals.
- Experiments with some small data sets are shown here.
Thanks! I've noticed that .RData files were a lot smaller size than csv.
ReplyDeleteHI Erwin, I came across this error when was trying to load .Rdata into RStudio. ‘file.rdata’ has magic number 'RDX3'
ReplyDeleteUse of save versions prior to 2 is deprecated. How do we deal with this?