While building a prototype .Rdata writer we developed a gdx2r application. This tool will convert a GAMS GDX file to an .RData file. Here we do some performance testing to get a feeling of how fast this is and how large the files are.
The first step is to generate a large, dense three dimensional parameter and export this to a GDX file. We can use compression to make the generated GDX file smaller at the expense of more CPU time.
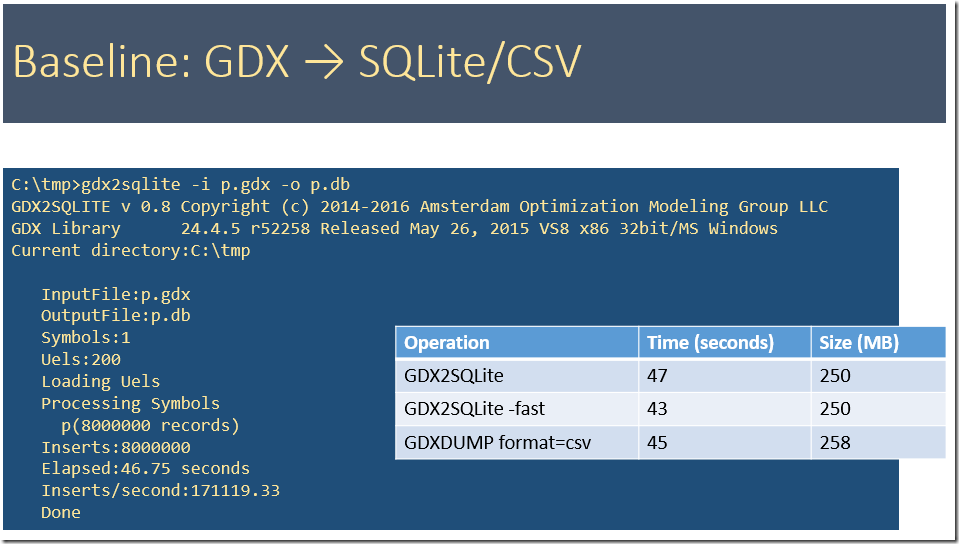
To get some baseline data we first see how our gdx2sqlite tool behaves. A SQLite database is a convenient format to get data into R or Python. The –fast option helps very little here. This option typically has large impact on datasets with many smaller tables. The SQLite database is in general larger than a GDX file. A big advantage of using a database format is that we can make queries in R or Python to import suitable subsets of the data. In addition these queries can do some operations (summarizing, pivoting, transforming etc.).
A second much used format is CSV files. The GDXDUMP tool can generate this file. R has a read.csv command to import CSV files. The CSV format is limited to a single table.
Below we see that we can generate large SQLite files very fast: comparable to writing a CSV files. Typically we don’t get these speeds when doing inserts into a server based RDBMS (such as SQL Server or Oracle). In the tlimings below we used the uncompressed GDX file.
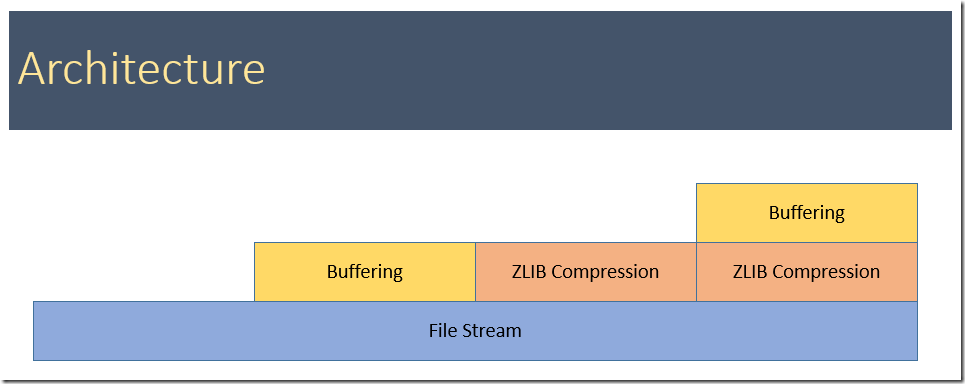
Below are timings from the gdx2r tool. We notice that buffering before passing data to the next layer (either the file stream or the compressed file stream) really improves the speed. Even a small buffer of 1000 bytes is doing a good job (from 0 to 1k bytes makes more of a difference than from 1k to 10k bytes). The explanation for this is the following: the calls to the file stream are passed on to the Windows API WriteFile and these calls have a large fixed cost. A similar argument holds for compressed file stream. Because of the different building blocks (buffering vs no buffering, compression vs no compression) we have to try a number of possibilities.
In the table below, when bufsize=0, no buffering is used.
Finally it is interesting to compare the reading times to get thigs into R. Below we have:
- Read table p from the SQLite database. This is relatively fast.
- Read the CSV file. The is very slow compared to the alternatives.
- Load the .RData file is also fast.
- The last entry is using the GAMS package gdxrrw. That package can read GDX files directly (assumes an installed GAMS system). This is somewhat slower than I would expect. It should be similar to SQLite and .Rdata timings but it is a factor of 2 slower.



No comments:
Post a Comment