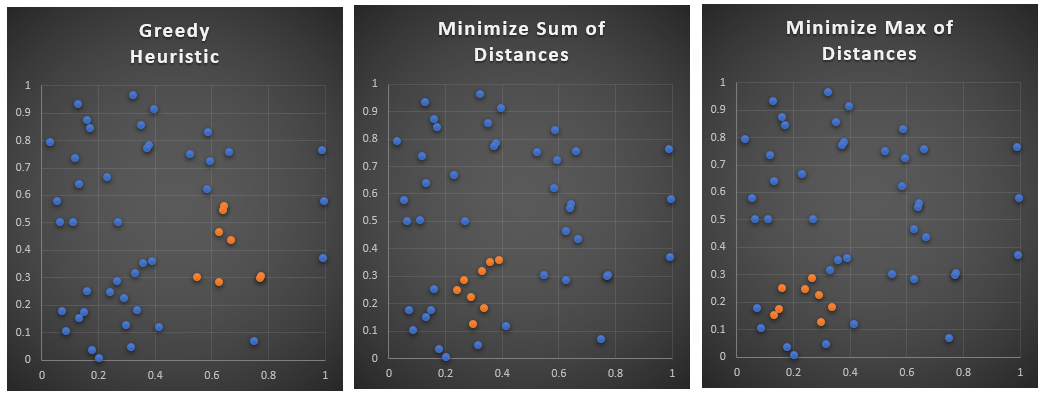
Problem statement
Consider \(n=100\) points in an 12 dimensional space. Find \(m=8\) points such that they are as close as possible.
Models
| MINSUM MIQP Model |
|---|
| \[\begin{align} \min & \sum_{i\lt j} \color{darkred}x_i \cdot \color{darkred}x_j \cdot \color{darkblue}{\mathit{dist}}_{i,j} \\ & \sum_i \color{darkred}x_i = \color{darkblue}m \\ & \color{darkred}x_i \in \{0,1\} \end{align}\] |
This is a simple model. The main wrinkle is that we want to make sure that we only count each distance once. For this reason, we only consider distances with \(i \lt j\). Of course, we can exploit this also in calculating the distance matrix \( \color{darkblue}{\mathit{dist}}_{i,j}\), and make this a strictly upper-triangular matrix. This saves time and memory.
| MINSUM MIP Model |
|---|
| \[\begin{align} \min & \sum_{i\lt j} \color{darkred}y_{i,j} \cdot \color{darkblue}{\mathit{dist}}_{i,j} \\ & \color{darkred}y_{i,j} \ge \color{darkred}x_i + \color{darkred}x_j - 1 && \forall i \lt j\\ & \sum_i \color{darkred}x_i = \color{darkblue}m \\ & \color{darkred}x_i \in \{0,1\} \\ &\color{darkred}y_{i,j} \in [0,1] \end{align}\] |
The inequality implements the implication \[\color{darkred}x_i= 1 \textbf{ and } \color{darkred}x_j = 1 \Rightarrow \color{darkred}y_{i,j} = 1 \] The variables \(\color{darkred}y_{i,j}\) can be binary or can be relaxed to be continuous between 0 and 1. The MIQP and MIP model should deliver the same results.
| MINMAX MIP Model |
|---|
| \[\begin{align} \min\> & \color{darkred}z \\ & \color{darkred}z \ge \color{darkred}y_{i,j} \cdot \color{darkblue}{\mathit{dist}}_{i,j} && \forall i \lt j \\ & \color{darkred}y_{i,j} \ge \color{darkred}x_i + \color{darkred}x_j - 1 && \forall i \lt j\\ & \sum_i \color{darkred}x_i = \color{darkblue}m \\ & \color{darkred}x_i \in \{0,1\} \\ &\color{darkred}y_{i,j} \in [0,1] \end{align}\] |
We re-use our linearization here.
- Select the two points that are closest to each other.
- Select a new unselected point that is closest to our already selected points.
- Repeat step 2 until we have selected 8 points.
Small data set
---- 10 PARAMETER coord coordinates x y point1 0.172 0.843 point2 0.550 0.301 point3 0.292 0.224 point4 0.350 0.856 point5 0.067 0.500 point6 0.998 0.579 point7 0.991 0.762 point8 0.131 0.640 point9 0.160 0.250 point10 0.669 0.435 point11 0.360 0.351 point12 0.131 0.150 point13 0.589 0.831 point14 0.231 0.666 point15 0.776 0.304 point16 0.110 0.502 point17 0.160 0.872 point18 0.265 0.286 point19 0.594 0.723 point20 0.628 0.464 point21 0.413 0.118 point22 0.314 0.047 point23 0.339 0.182 point24 0.646 0.561 point25 0.770 0.298 point26 0.661 0.756 point27 0.627 0.284 point28 0.086 0.103 point29 0.641 0.545 point30 0.032 0.792 point31 0.073 0.176 point32 0.526 0.750 point33 0.178 0.034 point34 0.585 0.621 point35 0.389 0.359 point36 0.243 0.246 point37 0.131 0.933 point38 0.380 0.783 point39 0.300 0.125 point40 0.749 0.069 point41 0.202 0.005 point42 0.270 0.500 point43 0.151 0.174 point44 0.331 0.317 point45 0.322 0.964 point46 0.994 0.370 point47 0.373 0.772 point48 0.397 0.913 point49 0.120 0.735 point50 0.055 0.576
HEURISTIC MIQP MIP MINMAX point2 1.000 point3 1.000 1.000 1.000 point9 1.000 point10 1.000 point11 1.000 1.000 point12 1.000 point15 1.000 point18 1.000 1.000 1.000 point20 1.000 point23 1.000 1.000 1.000 point24 1.000 point25 1.000 point27 1.000 point29 1.000 point35 1.000 1.000 point36 1.000 1.000 1.000 point39 1.000 1.000 1.000 point43 1.000 point44 1.000 1.000 status Optimal Optimal Optimal obj 3.447 3.447 0.210 sum 4.997 3.447 3.447 3.522 max 0.291 0.250 0.250 0.210 time 33.937 2.125 0.562
Large data set
---- 112 PARAMETER results HEURISTIC MIQP MIP MINMAX point5 1.000 1.000 point8 1.000 1.000 point17 1.000 1.000 point19 1.000 1.000 point24 1.000 point35 1.000 1.000 point38 1.000 1.000 point39 1.000 point42 1.000 1.000 point43 1.000 point45 1.000 1.000 1.000 point51 1.000 point56 1.000 point76 1.000 point81 1.000 1.000 point89 1.000 1.000 point94 1.000 1.000 1.000 1.000 point97 1.000 status TimeLimit Optimal Optimal obj 26.230 26.230 1.147 sum 30.731 26.230 26.230 27.621 max 1.586 1.357 1.357 1.147 time 1015.984 49.000 9.594 gap% 23.379
Conclusions
References
- Given a set of points or vectors, find the set of N points that are closest to each other, https://stackoverflow.com/questions/64542442/given-a-set-of-points-or-vectors-find-the-set-of-n-points-that-are-closest-to-e
Appendix: GAMS code
- There are not that many models where I can use xor operators. Here it is used in the Greedy Heuristic where we want to consider points \(i\) and \(j\) where one of them is in the cluster and one outside.
- Of course instead of the condition that x.l(i) xor x.l(j) is nonzero, we also could have required that x.l(i)+x.l(j)=1. I just could not resist the xor.
- Macros are used to prevent repetitive code in the reporting of results.
- Acronyms are used in reporting.
|
$ontext |